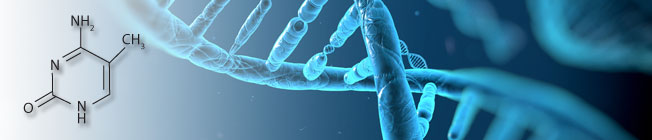
Global DNA Methylation Methods
Since the discovery of 5-methylcytosine (5-mC), researchers have been investigating the ubiquitous epigenetic modification known as DNA methylation. Measuring the total amount of 5-mC or 5-hmC (5-hydroxymethylcytosine) allows researchers to gain insight into profound biological processes and identify biomarkers for disease by, for example, comparing the DNA of normal versus diseased populations. Over the years we have seen the transformation of global DNA methylation assays available and the rise of new technological developments for accomplishing the quantitation of overall 5-mC content in DNA samples. Numerous methods and assays exist, each with their own benefits and limitations, limitations such as large input DNA requirements, sample species restrictions, extensive knowledge of complex protocols, or having to buy and set up costly equipment.
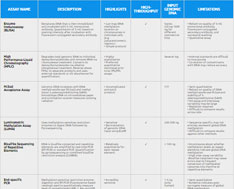
This comparison chart offers an overview of the advantages, disadvantages and capabilities of multiple global DNA methylation methods available, including HPLC, LC-MS/MS, LUMA, ELISA-based assays, and more. See how to overcome the limitations of various global DNA methylation assays.
| Assay Name | Description | Highlights | High-throughput | Input Genomic DNA | Limitations | |
|---|---|---|---|---|---|---|
| Enzyme Immunoassay (ELISA) | DNA is denatured and then immobilized and incubated with 5-mC monoclonal antibody. Quantitation is based on microplate spectrophotometer staining intensity reading after incubation with fluorescein-conjugated secondary antibody. | • Commercial kits available • Simple protocol • Can map DNA methylation location on chromosomes in individual cells (e.g. juxtacentromeric regions) | ✓ | Between 20-1000 ng for different commercial kits | • Reliant on quality of 5-mC monoclonal antibody, fluorescein-conjugated secondary antibody, and successful washing | Compare ELISA-based Global DNA Methylation Assay Kits |
| High Performance Liquid Chromatography (HPLC) | Total genomic DNA is degraded to individual deoxyribonucleotides and RNA is removed via ribonuclease treatment. Then converted to deoxyribonucleosides via alkaline phosphatase treatment. Reverse phase HPLC separates products and then external standards or UV absorbance are used for quantification. | • Highly quantitative • Preferred quantitative technique prior to LC-MS/MS assays | Several mg | • Internal standards are difficult to incorporate • Co-elution of contaminants with DNA may reduce accuracy | ||
| Liquid Chromatography-tandem Mass Spectrometry (LC-MS/MS) | Internal standards are spiked, but the procedure is the same as HPLC. Quantitation using mass spectrometry. | • Sensitive • Reproducible • Absolute quantification | ✓ | 100-500 ng | • Costly to set up instruments • Costly to make or buy internal standards • Expertise in complex mass spec procedure necessary | |
| M.SssI Acceptance Assay | Genomic DNA is incubated with DNA methyltransferase (M.Sssl) and methyl donor S-adenosylmethionine (SAM). DNA is then immobilized on nitrocellulose paper and scintillation counter then measures ionizing radiation. | • Uncomplicated and quick protocol | ✓ | • Semi-quantitative • Reliant on quality of DNA methyltransferase M.SssI and stability of S-adenosylmethionine (SAM) • Intrassay and interassay variability may be large • Radiation required • Difficult to compare results across labs | ||
| Luminometric Methylation Assay (LUMA) | Methylation-sensitive and insensitive restriction enzymes (HpaII, MspI, and EcoRI) are used to cleave DNA followed by polymerase extension assay by pyrosequencing. | • Sensitive • Strong internal control using EcoRI | ✓ | 200-500 ng | • Sequence specific; may not entirely represent global DNA methylation • Difficult to compare results against other methods | |
| Bisulfite Sequencing of Repetitive Elements | DNA is bisulfite-converted and repetitive elements are amplified by real-time PCR (RT-PCR) or standard PCR. Quantification by pyrosequencing or combined bisulfite restriction analysis (COBRA). | • Relatively quantitative for each repeat element | ✓ | < 100 ng | • Inconclusive about whether methylation levels at repeat elements indicate global DNA methylation • High inter-assay variability • Bisulfite treatment may cause errors due to the frequent conversion of methylated cytosines into thymine at repetitive elements | |
| End-specific PCR | Methylation-sensitive restriction enzyme digestion followed by RT-PCR and fluorescence-based readout are used to quantitatively measure level of unmethylated LINE-1, Alu and LTR. | • Incredibly sensitive | ✓ | 1-5 ng (only human) | • Semi-quantitative • Evidence indicates it may be a poor surrogate measure of global DNA methylation • Restricted to human DNA samples only • High background noise from highly heterogeneous regions • Can only assess certain subsets of LINE and Alu repeats |
Creating places that enhance the human experience.
Useful options
Beautiful snippets
Amazing pages
Building Castle in Cloud
When designing an architectural concept, we take into account the nine criteria that Zumthor considers to be important in creating an architectural atmosphere.